Service Project # 5: Paracrine interactions during cardiac and endothelial co-differentiation of HPSCS
Collaborating Investigator: Sean Palecek
Affiliation: University of Wisconsin – Madison
Funding Status: NIH R01EB007534
Project Period: 07/01/07-11/30/19
Summary
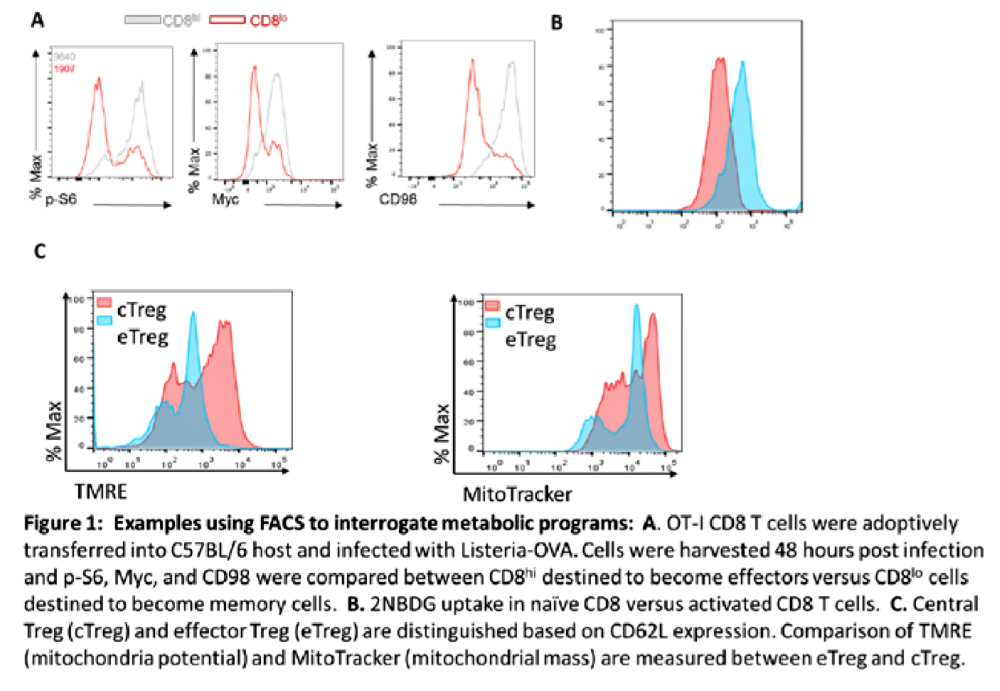
The overall goal of SP #6 is to develop a FACS-based system to interrogate metabolic status of cells. This SP will not develop any novel technology but rather push novel approaches to examining metabolic programming of cells using already technology. This FACS-based “individual” cell analysis will be developed (push) to interrogate cardiac HPSCS.
Specific Aim 1. Create standardized antibody panels and procedures for interrogating metabolic programing in cells using FACS.
Approach
As seen in Figure 1, our laboratory has been employing FACS to evaluate metabolic programming in T cells. In this proposed SP we will systematically create multi-antibody panels (upwards of 12 different antibodies at a time) to simultaneously measure metabolic signaling & function. We will standardize the panels for T cells making this technique operational for TRD 1 & 3 as well as our collaborators (Borrello and Sadelain). These panels will be expanded to diverse cell types for example iPSC (Palecek), Myocytes (Kass), Cancer cells (Zahnow) and Macrophages (Horton). Some examples of the work flow for SP6 can include: (1) Getting fresh (Fed-Ex Overnight) or fixed cells from investigator and providing data back, (2) Getting fresh or fixed cells from investigator, working out panels and getting initial results then sending procedure to investigator to pursue locally, and (3) Receiving fresh or fixed cells from investigator of cells genetically or chemically treated then screening cells for a desired metabolic effect.